A hierarchy of protein patterns robustly decodes cell shape information
Manon C. Wigbers, Tzer Han Tan, Fridtjof Brauns, Jinghui Liu, S. Zachary Swartz, Erwin Frey, Nikta Fakhri
In this work, we demonstrate how protein-based pattern formation in cells can be robust against large changes of cell shape. Specifically, we show how a surface contraction wave in starfish oocytes adapts to the cell shape to ensure that it robustly arrives at the same nucleus-located target position on the membrane, regardless of the cell shape. Using a combination of biophysical theory and experiments, we uncover a novel shape-adaptation mechanism that arises from a hierarchical sequence of protein patterns, i.e. by “patterns forming patterns” (Turing, 1952). Such a mechanism based on a hierarchy of patterns may constitute a general physical principle to transduce cell-shape information to biochemical regulators, facilitating cell-shape sensing, cell- shape control and protein positioning.
The interplay between protein patterns and cell mechanics is central to a long-standing puzzle, first formulated by Alan Turing in his seminal paper on “The chemical basis of morphogenesis”. There he notes that “the description of the state depends on two parts, the mechanical and chemical,” and that “the interdependence of the chemical and mechanical data adds enormously to the difficulty.” Thus, there are two distinct scenarios: the formation of functional protein patterns could either rely on the interplay with cell mechanics, or the formation of patterns has to adapt to mechanical influences in order to be robust.
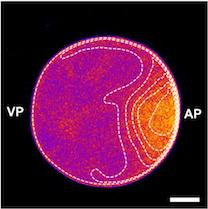
We studied how protein patterns robustly perform their specific biological function despite drastic cell shape deformations. Our experimental collaboration partners from MIT(Nikta Fakhri and her PhD student Tzer Han Tan) confined starfish oocytes in differently shaped, microfabricated chambers and imaged the spatiotemporal distribution of the cell shape regulator, RhoA. In starfish oocytes, just before meiotic cell division, RhoA travels as a pulse over the membrane, leading to a surface contraction wave and ultimately to polar body extrusion. Strikingly, as they forced these cells into different shapes, they found that despite dramatic changes to the propagation of the RhoA pulse, it always arrives exactly at its nucleus-located target position on the membrane. These intriguing observations show that the RhoA pulse ‘knows’ the cell shape and adapts to it.
Our group developed a biophysical theory that reveals that this shape-adaptation mechanism arises from a hierarchical coupling of protein patterns: The shape information is first encoded in a cytosolic gradient of the cell cycle regulator Cdk1, subsequently decoded by a bistable switch (RhoGEF Ect2), and finally transduced into a mechanochemical response via excitable dynamics of RhoA. Importantly, our work illustrates the importance of the self-organized formation of hierarchical patterns in understanding emergent biological functions, complementing genetic and molecular studies. Specifically, we would like to highlight two important implications: First, the Cdk1-Ect2- RhoA hierarchy establishes a mechano-chemical feedback loop, coupling shape information to a cell shape regulator. Thus, the Cdk1-Ect2-RhoA hierarchy may also be important for how cells control cell-shape.
Second, the Cdk1-Ect2-RhoA hierarchy integrates two main paradigms in the field of protein pattern formation: Turing’s idea that proteins self-organize from a homogeneous state into patterns via reaction and diffusion, and Wolpert’s idea that cells have a notion of positional information, which they use to form patterns. In our shape-adaptation mechanism, the Cdk1 gradient encodes positional information within the cell, which is subsequently transmitted via reaction-diffusion based 'protein modules’ (bistable Ect2 and excitable RhoA dynamics). These modules effectively perform computational operations (Ect2 measures a threshold and RhoA takes a temporal derivative) to process the positional information and ultimately lead to functional protein patterns. We believe that our mechanism can be generalized to a wide range of processes in which spatial information is processed, such as cell migration and cytokinesis. In particular, we are currently extending our biophysical theory to explain cleavage furrow positioning during cytokinesis in C. elegans.
In summary, we demonstrate a shape-adaptation mechanism, in which spatial information in cells of different shapes is processed by reaction–diffusion based protein modules. We envision that our results may provide new insights to other experimental model systems in which a precise spatial organization of proteins is essential.

