Nanopore translocation of structured RNA / DNA
Malcolm McCauley, Robert Forties, Ulrich Gerland and Ralf Bundschuh
Translocation through nanopores has emerged as a new experimental technique to probe the physical properties of biomolecules. The question of how the typical translocation time for a single unstructured polymer depends on its length has already triggered many theoretical and computational studies. We address this question for structured RNA molecules where the breaking of base-pairing patterns is the main barrier for translocation.
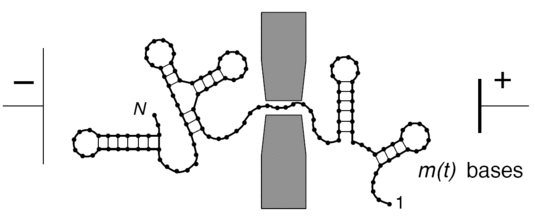
It is believed that with respect to these base-pairing patterns, a single-stranded RNA with a random sequence is typically either frozen into a single dominant structure (glassy phase) or many different structures with similar total energies coexist (molten phase), depending on the temperature and the base-pairing energetics. So far, these phases have not been confirmed experimentally. Our combined theoretical and computational study, based on a simple model for RNA folding and translocation, suggests that these two phases can be distinguished by their translocation behavior in the absence of an external voltage bias.

