Spontaneous Unknotting of a Polymer Confined in a Nanochannel
Wolfram Möbius, Erwin Frey, and Ulrich Gerland
Chip-based fluidic systems offer enormous prospects for analyzing, sorting, and manipulating complex molecules such as DNA and proteins. To this end, much fundamental research, both experimental and theoretical, is still required. We contribute a theoretical study on an obstacle for these applications: the formation of knots in long polymers, e.g. DNA, upon threading into narrow channels or pores. Such knots can hinder the transport of these chains, and they also undermine a key advantage of nano-confinement: only without knots does DNA in nanochannels take on a stretched-out conformation with a one-to-one correspondence between the position along the channel and the DNA sequence. This correspondence is crucial for mapping out local sequence-dependent properties, e.g. affinity for protein-binding.
Given that knots spontaneously arise in linear polymers, by which mechanism(s) do they dissolve under confinement in a channel, and how long does it take? And can one accelerate the process of unknotting externally? In our paper we address these questions using a multi-scale approach: we perform detailed simulations of the knotted polymer dynamics over a limited timescale, and show that the long-time behavior is in fact adequately described by three amazingly simple modes of motion, forming the basis for our coarse-grained theory of knot dynamics under confinement. We find that unknotting involves an interplay between diffusion of the knot along the polymer contour and variation of the knot size, leading to a characteristic dependence of knot lifetime on polymer length. Interestingly and quite counterintuitively, we find that one can accelerate unknotting considerably by exerting a weak pulling force, e.g. mediated by hydrodynamic drag.
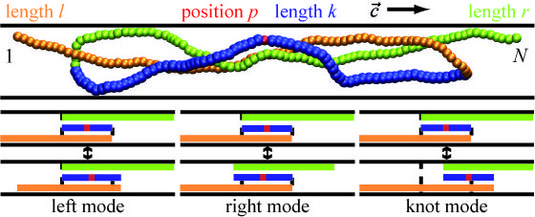
accepted for publication in Nano Letters

