Design of biochemical pattern forming systems from minimal motifs
Philipp Glock, Fridtjof Brauns, Jacob Halatek, Erwin Frey and Petra Schwille
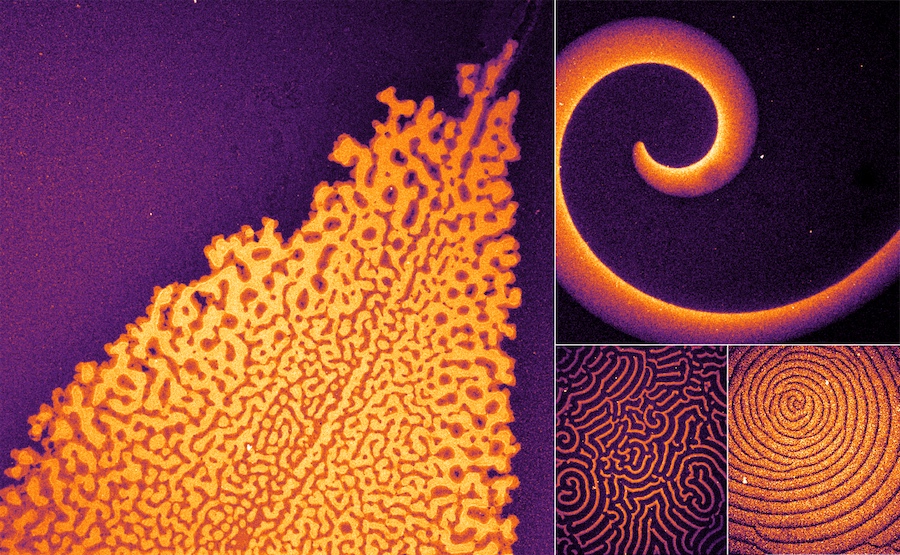
Self-organized concentration patterns are crucial for the development and function of organisms on all levels. However, there is a large gap of knowledge on how different molecular features determine the emergence of patterns. Systematic design on the molecular level requires understanding of the underlying principles and the role of specific molecular features.
In this paper, we approach the task of modular engineering of protein pattern formation in a novel approach, based on our combined experience in both, in vitro reconstitution and in silico theoretical analysis of pattern-forming protein networks.
Using the two-protein MinDE system from Escherichia coli as a starting point, we reduce the catalytic unit of MinE to a minimal, 19 amino acid peptide. This catalytic unit is unable to form patterns with MinD by itself, but pattern formation can be rescued in combination with either an artificial dimerization or membrane-targeting module, or both. Thus, we supply a modular reaction-diffusion toolkit in which the individual functions of our MinE substitute can be directly related to specific outcomes of the self-organization assay. Furthermore, analysis of our mathematical model reveals that the mechanisms between recombination-driven pattern formation and recruitment-driven pattern formation are different and that in vivo, the recruitment-driven mechanism is likely dominant.

