Rationally designed logic signal integration in mammalian cells
M. Leisner, L. Bleris, J. Lohmueller, Z. Xie, K. Benenson
Experiments performed and published with Benenson lab, Harvard
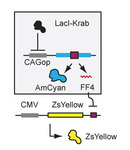 The capacity to reprogram and control cell functions in a specified manner is needed in many areas of biotechnology, bioengineering and medicine. Biological “computers”, or “automata” have been proposed as a way to achieve this capacity. These systems are modular biological networks that in principle should be able to convert an arbitrary specified set of biological input signals into specified responses (output) according to a desired set of rules, or program. Challenging aspects hereby are modularity, programmability as well as scalability of a given system.
The capacity to reprogram and control cell functions in a specified manner is needed in many areas of biotechnology, bioengineering and medicine. Biological “computers”, or “automata” have been proposed as a way to achieve this capacity. These systems are modular biological networks that in principle should be able to convert an arbitrary specified set of biological input signals into specified responses (output) according to a desired set of rules, or program. Challenging aspects hereby are modularity, programmability as well as scalability of a given system.
Natural regulatory networks, in particular in mammalian cells, operate on many levels, including long-term epigenetic modifications, chromatic structure, availability of transcription factors, RNA splicing and transport, translation, post-translational modifications and localization. While all these mechanisms are candidates for forward engineering of regulatory functions, in practice only a few mechanisms are sufficiently well-studied to enable their rational manipulation. These include control of mRNA levels by trans-factors such as transcription factors (TFs), small RNAs in RNA interference pathway, and cis-acting riboswitches. Here we propose and support by experiments an approach towards implementing an arbitrarily defined gene regulatory function using transcription factor inputs and microRNA intermediates acting on response elements in the output gene.

